Catégorie d'actualités : PASTEUR
« Les métaux et la vie », « Pasteur au microscope » : représentations théâtrales à venir
La chimie s’invite au théâtre ! Venez découvrir :
- « Les métaux et la vie » à la Biennale du vivant
Samedi 27/09 à 18h30 – théâtre Nicole Loraux (45 rue d’Ulm)
Événement gratuit sur réservation : https://www.eventbrite.fr/e/billets-biennale-du-vivant-jour-2-ecole-normale-superieure-psl-1510078057119 - « Pasteur au microscope » à la Fête de la science
Dimanche 5/10 à 16h – salle Dussane (45 rue d’Ulm)
Événement gratuit sur réservation : https://www.eventbrite.fr/e/billets-fete-de-la-science-1664207763219?aff=oddtdtcreator
Une belle occasion de découvrir que nous serions ni vivants, ni humains, sans les métaux, et la vie de Pasteur à quelques mètres de son ancien bureau à l’ENS !
Synthetic cells: poles in good position!
Living systems, such as cells, are characterized by a precise spatio-temporal distribution of their components. In order to understand the mechanisms that govern live cell functions, scientists sometimes build simplified models of cells, commonly known as synthetic cells or artificial cells. These systems, with a perfectly well-known composition and on which one can act at will, make it possible to study a variety of phenomena and understand certain essential elements. However, to date, most synthetic cells are globally isotropic objects, with no top or bottom, no left or right, no head or feet – a far cry from the multi-component, polarized structures of living cells.
We therefore started with one of the most widely-used cellular models, the giant unilamellar vesicle (GUV), and developed a method for not only polarizing it, but also controlling the number of its poles and positioning them in a user-defined manner. A GUV is a closed lipid bilayer separating an internal aqueous medium from another outer aqueous medium. Generally spherical in shape when unconstrainted, its size varies from a few micrometers to a few hundred micrometers, making it comparable to cellular dimensions. For this work, we used membranes composed of lipid mixtures leading to the coexistence of two phases: an liquid-ordered phase (Lo) and a liquid-disordered phase (Ld). If the Lo phase dominates, Ld domains are formed within the continuous Lo phase, and vice versa. This well-known phenomenon is often used to mimic domain formation in biological membranes. However, these domains are mobile, impossible to position, and tend to merge to minimize the energy of the contact line between the two phases. This results in a single, randomly positioned domain composed of the minority phase.

Figure 1. By confining giant unilamellar vesicles (GUVs) in different geometries, it is possible to position lipid domains deterministically, enabling the creation of cellular models with 2, 3 or 4 poles. In these images, the liquid-disordered (Ld) phase appears in red, the liquid-ordered (Lo) phase in blue.
To guide the organization and localization of these domains, we confined the GUVs in order to deform them and generate, within each GUV, some free curved zones so as to promote the fusion of minority-phase domains by minimizing line energy. We first confined the GUVs in microchannels and showed that the minority-phase domains accumulate systematically at the free curved ends and merge there to form a single domain at each extremity, thus creating a GUV with two “poles” (Figure 1, left). Interestingly, it’s not the chemical nature of the lipid that determines its position, but rather the fraction of its phase. It is thus possible to create Ld or Lo poles, yet having very different lipid compositions, simply by adjusting the respective fraction of the two phases. Using a variety of confinement geometries, including pillar arrays, we were finally able to create GUVs with 3 or 4 poles (Figure 1, middle and right) positioned in a deterministic manner.
This work shows for the first time how to control the number and position of poles, in the form of lipid domains, within GUVs used as simplified cell models. By coupling the position of these domains with other components (DNA, proteins), this method will make it possible to control or study, within model systems, the spatio-temporal organization of key elements involved in a variety of processes such as cytoskeletal dynamics, cell division, regulatory processes or compartmentalization. This work also highlights the fundamental role of line energy in self-organization phenomena within biological or reconstituted membranes.
Reference : K. Nakazawa, A. Lévrier, S. Rudiuk, A. Yamada, M. Morel, D. Baigl.* Controlled Lipid Domain Positioning and Polarization in Confined Minimal Cell Models. Angew. Chem. Int. Ed. 2025, e202419529
Link (open access) : https://doi.org/10.1002/anie.202419529
Contact : Damien Baigl (damien.baigl@ens.psl.eu)
Les mécanismes de réactions clés des origines de la vie révélés par des simulations moléculaires guidées par l’intelligence artificielle
Et si nous pouvions utiliser l’intelligence artificielle pour démêler tous les détails moléculaires complexes des réactions chimiques, explorer des voies de réaction distinctes, révéler la participation des molécules de solvant et prendre en compte toutes les nuances d’effets thermodynamiques ? C’est le tour de force qu’ont réalisé des chercheurs du laboratoire PASTEUR (CNRS/ENS Paris/Paris Sciences et Lettres/Sorbonne Université) dans deux publications sorties dans les Proceedings of the National Academy of Sciences USA et le Journal of the American Chemical Society. En proposant une méthode systématique pour entraîner des neurones artificiels capables de s’affranchir des limitations actuelles des simulations moléculaires au niveau quantique, ils ont jeté un œil nouveau sur deux réactions cruciales de la chimie prébiotique, à l’origine de la synthèse des protéines et des acides nucléiques. En identifiant les mécanismes les plus favorables pour ces deux réactions, qui échappaient encore à l’expérience, ces résultats ouvrent la voie à la compréhension de la façon dont ces réactions ont pu se produire dans un contexte où les catalyseurs du vivant (les enzymes) n’existaient pas encore.
Les réactions chimiques sont des événements rares dans la vie des molécules. Observer comment les molécules réagissent est un défi expérimental ; même avec un microscope géant, nous passerions la majeure partie de votre temps à attendre qu’elles réagissent, et quand elles le font, cela se produit si rapidement que nous le manquerions. C’est là que les simulations entrent en jeu: elles permettent de suivre ces événements fugaces à notre guise. Mais elles rencontrent deux problèmes. Premièrement, suivre des événements réactifs signifie résoudre l’équation de Schrödinger des milliers de fois, ce qui est notoirement difficile. Deuxièmement, même après un effort de calcul aussi immense, nous n’aurions qu’une poignée d’observations. Mais la physique statistique nous enseigne que les systèmes atomistiques sont probabilistes, et des centaines d’observations sont nécessaires pour obtenir une image correcte et des quantités thermodynamiquement significatives.
Dans deux études séparées à la méthodologie voisine, les chimistes de l’ENS ont utilisé une approche d’intelligence artificielle émergente pour surmonter ces limitations. L’idée est d’entraîner un réseau de neurones à résoudre l’équation de Schrödinger pour toutes les structures possibles rencontrées lors d’une réaction chimique donnée. Un élément critique pour le succès de la démarche est la curation des données, pour laquelle ils ont proposé une méthode standardisée et robuste. Le résultat ? Une approche qui comble le fossé entre les mondes de la physique quantique et de la physique statistique à un coût de calcul modeste (et donc une empreinte carbone faible). Le meilleur des deux mondes !
Ces travaux ouvrent la voie à une modélisation beaucoup plus abordable des réactions chimiques en phase condensée, et à un changement de paradigme quant à la complémentarité qu’elles peuvent apporter aux résultats expérimentaux parfois difficiles à obtenir. Appliquée à deux réactions fondamentales de la synthèse des protéines d’une part (la liaison peptidique) et des acides nucléiques d’autre part (la liaison phosphoester), cette méthode a permis de mettre en évidence les voies de réaction les plus favorables pour ces deux réactions, ce que ne permettaient pas directement les études expérimentales. Ces résultats s’affichent comme une première étape vers la compréhension des conditions physico-chimiques optimales pour ces réactions énergétiquement très défavorisées et très peu probables qui ont mené à la formation des premières briques élémentaires du vivant il y a un peu plus de quatre milliards d’années.
Références :
Prebiotic chemical reactivity in solution with quantum accuracy and microsecond sampling using neural network potentials, Z. Benayad, R. David, G. Stirnemann, Proceedings of the National Academy of Sciences 121, e2322040121 (2024)
Competing reaction mechanisms of peptide bond formation in water revealed by deep potential molecular dynamics and path sampling, R. David, I. Tuñón, D. Laage, Journal of the American Chemical Society 146, 14213-14224 (2024)
Contacts :
Guillaume Stirnemann – guillaume.stirnemann@ens.psl.eu
Damien Laage – damien.laage@ens.psl.eu
Département de Chimie, UMR 8640 PASTEUR, École normale supérieure, Paris
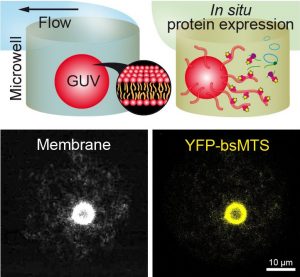
Sedimentation-Based Confinement of Individual Giant Unilamellar Vesicles in Microchamber Arrays with a Dynamically Exchangeable Outer Medium
by Syed Kaabir Ali, Catherine Sella, Vadim Dilhas, Barbara Jacková, Marina Mariconti, Clara Gomez-Cruz, Laurent Thouin, Mathieu Morel, Damien Baigl, and Ayako Yamada*
La fluorescence pour mesurer l’intensité lumineuse
English version below___“Fluorescence for measuring light intensity”
__Les mesures absolues d’intensité lumineuse sur une large gamme de longueurs d’onde et d’intensités lumineuses sont indispensables dans de nombreux domaines (bio-imagerie, optogénétique, photoproduction de molécules et de matériaux, conception de protocoles médicaux, photocatalyse…). Elles permettent en particulier de choisir rationnellement des paramètres tels que la durée d’application de la lumière, pour fournir le bon nombre de photons à un échantillon. Elles permettent également d’établir une comparaison équitable des résultats obtenus par différents groupes de recherche en garantissant ainsi la reproductibilité.Les protocoles actuellement disponibles aux biologistes, chimistes, ingénieurs et physiciens nécessitent un équipement et une expertise spécifiques. Ils souffrent aussi de limitations dans leur portée. Il existait ainsi un besoin de protocoles simples et courts pour accéder de manière robuste à cette mesure, une conclusion résonnant d’ailleurs avec les besoins soulevés par la communauté QUAREP-LiMi pour calibrer l’illumination dans des instruments optiques (https://quarep.org/).Cet article répond à ce besoin en rapportant deux protocoles qui exploitent la fluorescence pour permettre la mesure simple, rapide, et peu coûteuse de l’intensité lumineuse sur de larges gammes de longueurs d’onde et d’intensités, en fournissant des informations sur la distribution spatiale, y compris dans la profondeur des échantillons. Le premier protocole repose sur des espèces dont l’intensité de fluorescence diminue/augmente de manière mono-exponentielle lorsqu’une lumière constante est appliquée. Le second protocole fait appel à un fluorophore inerte photochimiquement à large absorption pour recalculer l’intensité lumineuse d’une longueur d’onde à l’autre. Pour démontrer leur utilité, ces protocoles ont été appliqués pour caractériser quantitativement la distribution spatiale de la lumière de divers systèmes d’imagerie par fluorescence et pour calibrer l’éclairage d’instruments et de sources lumineuses disponibles dans le commerce.
__Absolute measurements of light intensity over a wide range of wavelengths and light intensities are essential in many fields (bio-imaging, optogenetics, photoproduction of molecules and materials, design of medical protocols, photocatalysis, etc.). In particular, they enable a rational choice to be made of parameters such as the duration of light application, in order to deliver the right number of photons to a sample. They also enable a fair comparison of the results obtained by different research groups, thereby guaranteeing reproducibility.The protocols currently available to biologists, chemists, engineers and physicists require specific equipment and expertise. They also suffer from limitations in their scope. There was therefore a need for simple, short protocols for robustly accessing this measurement, a conclusion that resonates with the needs raised by the QUAREP-LiMi community for calibrating illumination in optical instruments (https://quarep.org/).This article responds to this need by reporting two protocols that exploit fluorescence to enable simple, rapid and inexpensive measurement of light intensity over wide ranges of wavelengths and intensities, providing information on spatial distribution, including in the depth of samples. The first protocol is based on species whose fluorescence intensity decreases/increases mono-exponentially when constant light is applied. The second protocol uses a photochemically inert fluorophore with broad absorption to recalculate light intensity from one wavelength to another. To demonstrate their usefulness, these protocols have been applied to quantitatively characterize the spatial light distribution of various fluorescence imaging systems and to calibrate the illumination of commercially available instruments and light sources.
– Lien(s) utile(s) : https://www.nature.com/articles/s41592-023-02063-y
.
Auteur(s)
Prénom(s) Nom(s) : Ludovic Jullien
Nom du laboratoire : PASTEUR UMR 8640
Tutelles : CNRS-ENS-SU
Informations édition
Revue : Nature Methods
Date de publication : 23/11/2023
Visuel
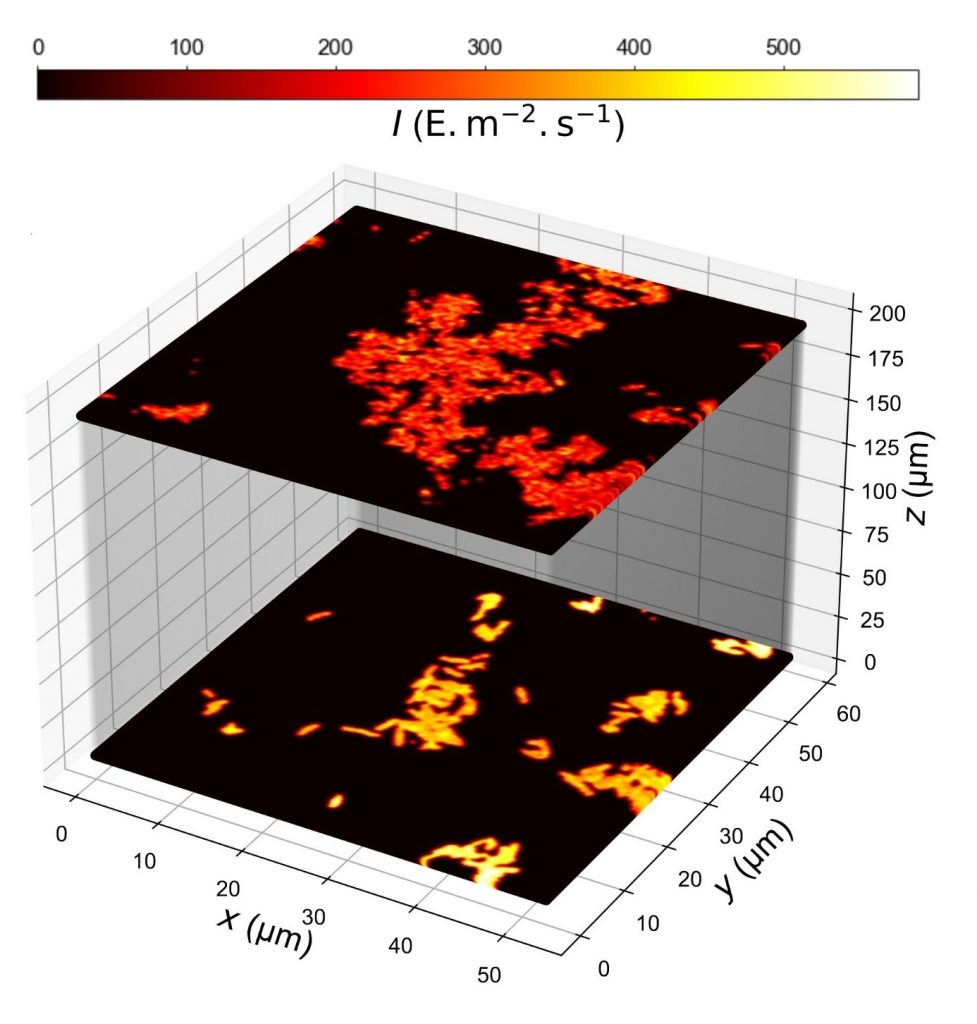
■ Légende : Map of light intensity in confocal microscopy retrieved from Dronpa-2-labeled E. Coli bacteria imaged at the surface or through a 2 % agarose pad by changing the sample orientation
■ Crédit : A. Lahlou et al, Nature Methods, 2023.
Fluorescent proteins glow longer in the presence of infrared light
Fluorescence imaging is widely used in the life sciences to visualize and analyze biological structures and processes. One of the main limitations is the photobleaching of fluorophores, i.e. their gradual photodegradation by the light used to excite fluorescence. Fluorophore photobleaching limits the duration and time resolution of experiments and degrades image quality. It also generates reactive oxygen species (ROS), which are toxic to living samples. Currently, two main approaches are used to control photobleaching. The first is to reduce the dose of excitation light, but this is usually at the expense of signal-to-noise ratio and/or time resolution. The second approach is to modify the fluorophore environment, generally to make it more reducing (removal of oxygen, addition of anti-oxidants) in order to neutralize ROS. However, this second approach is only of limited effectiveness, and it can disrupt the physiology of the biological systems of interest.
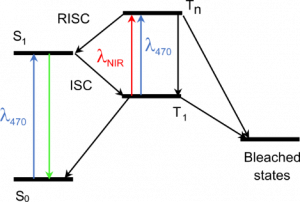
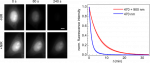
Principle of our method. The triplet excited state T1, a precursor of bleached states, is formed under visible excitation of the fluorophore from the bright singlet excited state S1. Near-infrared co-illumination promotes T1 into the higher triplet state Tn, from which the fluorophore returns to S1 by RISC.
In a paper published this month in Nature Biotechnology (https://doi.org/10.1038/s41587-023-01893-7), we describe a new method to reduce the photobleaching of fluorescent proteins and the associated phototoxicity. Our method exploits a photophysical process known as reverse intersystem crossing (RISC) to depopulate the triplet excited state of fluorophores, a precursor of photobleaching and a source of ROS. We induce RISC by co-illuminating fluorescent proteins at a near-infrared (NIR) wavelength absorbed by their triplet excited state, during their excitation with visible light. In our paper, we successfully apply this dual illumination method to wide-field fluorescence imaging of live eukaryotic and prokaryotic cells labelled with a wide range of green and yellow fluorescent proteins. We achieve a typical reduction in photobleaching by a factor of ~4, greater than that obtained with conventional antifading media. We show that our method can, for example, substantially improve the tracking of fluorescently-labelled replisomes in live E. coli cells. Finally, we show that it also reduces phototoxicity due to fluorescent protein excitation in growing bacteria and primary mouse neutrophils.
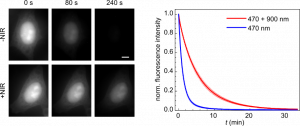
NIR co-illumination reduces the photobleaching of EGFP-labelled HeLa cells. Left: Fluorescence images of fixed HeLa cells labelled with EGFP and continuously illuminated at 470 nm alone (top) or combined with 900 nm (bottom). I470 = 32 W/cm2. I900 = 2 kW/cm2. Scale bar, 10 μm. Right: Photobleaching kinetics of HeLa cells. Values are mean ± s.d. (n = 12 cells).
Our method is efficient, technically easy to implement and compatible with living samples. It should therefore be widely adopted and provide access to a variety of new biological information, by allowing the use of higher light doses without adverse effects. Our work is also pioneering the use of RISC in fluorescence imaging, and can be expected to spark more applications in the near future, for example to other fluorophores or other imaging techniques. It should therefore have a major impact in the field.
Contacts: Agathe Espagne (agathe.espagne@ens.psl.eu) and Thomas Le Saux (thomas.lesaux@ens.psl.eu)
This work was carried out in collaboration with Lydia Robert (Micalis Institute, INRAE, Jouy-en-Josas, France).
Reference of the publication:
L. Ludvikova, E. Simon, M. Deygas, T. Panier, M.-A. Plamont, J. Ollion, A. Tebo, M. Piel, L. Jullien, L. Robert*, T. Le Saux* & A. Espagne*. Near-infrared co-illumination of fluorescent proteins reduces photobleaching and phototoxicity. Nat. Biotechnol. (2023). https://doi.org/10.1038/s41587-023-01893-7
Comment les bactéries aident à nettoyer les marées noires
Afin de mieux assainir les environnements pollués par d’importantes marées noires, des chercheurs ont étudié une bactérie marine mangeuse d’hydrocarbures. Cet organisme inhabituel, dont le nom scientifique est Alcanivorax borkumensis (Alca), peut survivre dans l’océan grâce à la seule consommation d’hydrocarbures. Des études récentes montrent que pratiquement toutes les bactéries forment des biofilms – des communautés tridimensionnelles maintenues ensemble par des substances extracellulaires – et, dans le cas d’Alca, qu’elles adhèrent à la surface des gouttelettes de pétrole.
Leurs biofilms déforment les gouttes de pétrole et mangent les hydrocarbures, ou ils peuvent parfois transformer les gouttes en structures agrégées plus grandes qui peuvent ensuite couler au fond de la mer. C’est ce qui est arrivé à une partie du pétrole de la marée noire de Deepwater Horizon en 2010. Deux questions essentielles se posent toutefois : comment les biofilms se forment-ils et leur dynamique influe-t-elle sur la consommation de pétrole ?
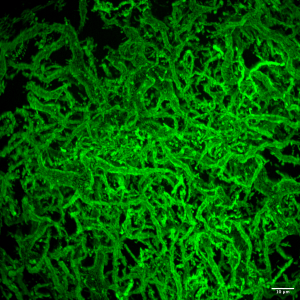
Source : Prasad et al. Science (2023)
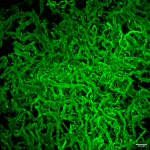
Andrew Utada (Université de Tsukuba), Jacques Fattaccioli (ENS-PSL, Sorbonne Université, IPGG), Jean-François Rupprecht (Aix-Marseille Université) et leurs collègues ont maintenant identifié le mécanisme de formation à l’aide d’un dispositif microfluidique qu’ils ont mis au point. En temps réel, ce dispositif permet de piéger et d’imager des gouttes d’huile chargées de bactéries. Grâce à ce dispositif, l’équipe a pu observer et quantifier l’ensemble du processus de formation du biofilm, depuis le début avec 20-50 cellules jusqu’à la consommation complète des gouttes d’huile.
L’équipe a observé deux morphologies de biofilms : les bactéries nourries pendant un jour ou moins formaient généralement des biofilms sphériques, tandis que celles qui suivaient un régime de cinq jours formaient une tête de méduse composée de tubes de biofilm minces et ramifiés, ou dendrites. De manière inattendue, les bactéries nourries pendant 2 à 4 jours ont alterné plus de 10 fois entre une forme sphérique et de grandes structures tubulaires (voir ci-dessus).
Les bactéries mangeuses de pétrole sécrètent des molécules de biosurfactant qui possèdent des régions hydrophiles et hydrophobes. Cette double propriété permet aux molécules de s’agencer sur les interfaces huile-eau afin de réduire la tension de surface, ce qui, selon Utada et ses collègues, permet à Alca de déformer plus facilement les gouttes d’huile pour leur donner une forme dendritique.. En déformant la goutte d’huile, les bactéries augmentent la surface de l’huile et accélèrent ainsi sa consommation. Les bactéries à la morphologie de biofilm sphérique ont consommé 90 % du volume d’huile après environ 72 heures, alors qu’il n’a fallu que 20 heures aux bactéries à la morphologie dendritique pour consommer le même volume.
Lien vers l’article :
Alcanivorax borkumensis Biofilms Enhance Oil Degradation By Interfacial Tubulation
Manoj Prasad, Nozomu Obana, S.-Z. Lin, Ken Sakai, Carles Blanch-Mercader, Jacques Prost, Nakao Nomura, Jean-François Rupprecht, Jacques Fattaccioli, A. S. Utada,
Science 381, 748-753 (2023)
Doi : 10.1126/science.adf3345
Lien vers l’article Perspective :
Bacteria stretch and bend oil to feed their appetite
T. J. McGenity, P. P. Laissue
Science 381, 728-729 (2023).
DOI:10.1126/science.adj4430
Contact: Jacques Fattaccioli (jacques.fattaccioli@ens.psl.eu)
Evolutive DNA origamis
Evolutive DNA origamis
The emergence of structural DNA nanotechnology has revolutionized nanoscience, by making it possible to program the assembly of large amounts of synthetic DNA bricks into virtually any desired morphology, as well as to position guests (molecules, proteins, nanoparticles) on such scaffolds with exquisite resolution. The resulting structures, and especially so-called DNA origamis, are currently used in a wide range of fundamental studies and applications in fields as various as chemistry, materials science, physics, biology and medicine. However, these structures are generally obtained by thermal annealing, in which the system is heated to a high temperature (around 80 degrees) prior to a slow cooling down process. The resulting objects are also essentially static and major reconfigurations are difficult to achieve.
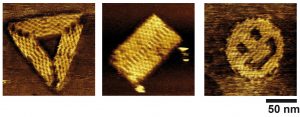
Examples of DNA origamis obtained by isothermal self-assembly at room temperature.
In an article published online this week (link : https://www.nature.com/articles/s41565-023-01468-2), we describe an innovative method in which hundreds of different DNA bricks can spontaneously self-assemble, at constant temperature (notably ambient, or physiological), into a desired shape. The method is generic and can be applied for the isothermal self-assembly of various kinds of structures (DNA origamis, nanogrids, self-assembled single-stranded tiles, etc) in both two or three dimensions, and allows the concomitant incorporation of proteins at prescribed positions. Using high-speed, high-resolution atomic force microscopy (AFM), we were able to image the assembly mechanism in situ and in real time for the first time, allowing us to reveal that the assembly processes through multiple folding pathways, the system escaping kinetic traps until finding its most stable, equilibrium shape. The resulting structures have also a remarkable reconfigurable and evolutive character. We show for instance that the system is capable to self-select its most stable shape in a complex mixture of competitive strands. Additionally, DNA origamis obtained by this method can spontaneously evolve each time a new energy minimum appears. We observed in particular the first origamis capable to isothermally shift from one initially stable shape (here, a rectangle) to a radically different morphology (here, a triangle) though a massive exchange of their constitutive DNA strands.
These results significantly enhance the prospects offered by isothermal self-assembly, making it possible to obtain sophisticated, programmable nanostructures at room or physiological temperature and allows to envision synthetic nanostructures capable of self-selection, adaptation and evolution.
The article (open access) can be freely downloaded from this link: : https://www.nature.com/articles/s41565-023-01468-2
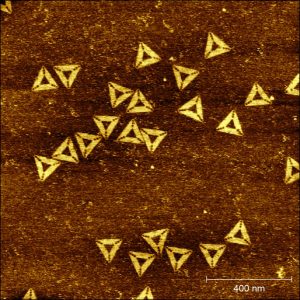
The method allows the spontaneous evolution of origamis from one shape to another one upon appearance of a new energy minimum. In this example, origamis having initially a rectangular shape, have evolved to perfectly well formed triangles.
Article information:
Title: Isothermal self-assembly of multicomponent and evolutive DNA nanostructures
Abstract: Thermal annealing is usually needed to direct the assembly of multiple complementary DNA strands into desired entities. We show that, with a magnesium-free buffer containing NaCl, complex cocktails of DNA strands and proteins can self-assemble isothermally, at room or physiological temperature, into user-defined nanostructures, such as DNA origamis, single-stranded tile assemblies and nanogrids. In situ, time-resolved observation reveals that this self-assembly is thermodynamically controlled, proceeds through multiple folding pathways and leads to highly reconfigurable nanostructures. It allows a given system to self-select its most stable shape in a large pool of competitive DNA strands. Strikingly, upon the appearance of a new energy minimum, DNA origamis isothermally shift from one initially stable shape to a radically different one, by massive exchange of their constitutive staple strands. This method expands the repertoire of shapes and functions attainable by isothermal self-assembly and creates a basis for adaptive nanomachines and nanostructure discovery by evolution.
Reference: C. Rossi-Gendron,§ F. El Fakih, § L. Bourdon, K. Nakazawa, J. Finkel, N. Triomphe, L. Chocron, M. Endo, H. Sugiyama, G. Bellot, M. Morel, S. Rudiuk & D. Baigl*. Isothermal self-assembly of multicomponent and evolutive DNA nanostructures. Nat. Nanotechnol. (2023). https://doi.org/10.1038/s41565-023-01468-2
§: equal contribution ; * : correspondence to: damien.baigl@ens.psl.eu
Contact: Damien Baigl (damien.baigl@ens.psl.eu)
Aerosol Jet-Printed High-Aspect Ratio Micro-Needle Electrode Arrays Applied for Human Cerebral Organoids and 3D Neurospheroid Networks
Aerosol Jet-Printed High-Aspect Ratio Micro-Needle Electrode Arrays Applied for Human Cerebral Organoids and 3D Neurospheroid Networks
Auteurs :
Sabine Zips, Boxin Huang, Salammbô Hotte, Lukas Hiendlmeier, Chen Wang, Karthyayani Rajamani, Olivier Buriez, George Al Boustani, Yong Chen, Bernhard Wolfrum, and Ayako Yamada
Abstract :
The human brain is a complex and poorly accessible organ. Thus, new tools are required for studying the neural function in a controllable environment that preserves multicellular interaction and neuronal wiring. In particular, high-throughput methods that alleviate the need for animal experiments are essential for future studies. Recent developments of induced pluripotent stem cell technologies have enabled in vitro modeling of the human brain by creating three-dimensional brain tissue mimic structures. To leverage these new technologies, a systematic and versatile approach for evaluating neuronal activity at larger tissue depths within the regime of tens to hundreds of micrometers is required. Here, we present an aerosol-jet- and inkjet-printing-based method to fabricate microelectrode arrays, equipped with high-aspect ratio μ-needle electrodes that penetrate 3D neural network assemblies. The arrays have been successfully applied for electrophysiological recordings on interconnected neurospheroids formed on an engineered substrate and on cerebral organoids, both derived from human induced pluripotent stem cells.