
Evolutive DNA origamis
The emergence of structural DNA nanotechnology has revolutionized nanoscience, by making it possible to program the assembly of large amounts of synthetic DNA bricks into virtually any desired morphology, as well as to position guests (molecules, proteins, nanoparticles) on such scaffolds with exquisite resolution. The resulting structures, and especially so-called DNA origamis, are currently used in a wide range of fundamental studies and applications in fields as various as chemistry, materials science, physics, biology and medicine. However, these structures are generally obtained by thermal annealing, in which the system is heated to a high temperature (around 80 degrees) prior to a slow cooling down process. The resulting objects are also essentially static and major reconfigurations are difficult to achieve.
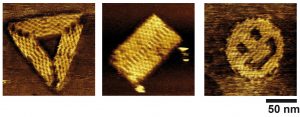
Examples of DNA origamis obtained by isothermal self-assembly at room temperature.
In an article published online this week (link : https://www.nature.com/articles/s41565-023-01468-2), we describe an innovative method in which hundreds of different DNA bricks can spontaneously self-assemble, at constant temperature (notably ambient, or physiological), into a desired shape. The method is generic and can be applied for the isothermal self-assembly of various kinds of structures (DNA origamis, nanogrids, self-assembled single-stranded tiles, etc) in both two or three dimensions, and allows the concomitant incorporation of proteins at prescribed positions. Using high-speed, high-resolution atomic force microscopy (AFM), we were able to image the assembly mechanism in situ and in real time for the first time, allowing us to reveal that the assembly processes through multiple folding pathways, the system escaping kinetic traps until finding its most stable, equilibrium shape. The resulting structures have also a remarkable reconfigurable and evolutive character. We show for instance that the system is capable to self-select its most stable shape in a complex mixture of competitive strands. Additionally, DNA origamis obtained by this method can spontaneously evolve each time a new energy minimum appears. We observed in particular the first origamis capable to isothermally shift from one initially stable shape (here, a rectangle) to a radically different morphology (here, a triangle) though a massive exchange of their constitutive DNA strands.
These results significantly enhance the prospects offered by isothermal self-assembly, making it possible to obtain sophisticated, programmable nanostructures at room or physiological temperature and allows to envision synthetic nanostructures capable of self-selection, adaptation and evolution.
The article (open access) can be freely downloaded from this link: : https://www.nature.com/articles/s41565-023-01468-2
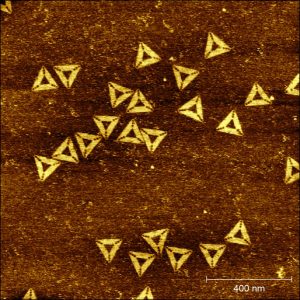
The method allows the spontaneous evolution of origamis from one shape to another one upon appearance of a new energy minimum. In this example, origamis having initially a rectangular shape, have evolved to perfectly well formed triangles.
Article information:
Title: Isothermal self-assembly of multicomponent and evolutive DNA nanostructures
Abstract: Thermal annealing is usually needed to direct the assembly of multiple complementary DNA strands into desired entities. We show that, with a magnesium-free buffer containing NaCl, complex cocktails of DNA strands and proteins can self-assemble isothermally, at room or physiological temperature, into user-defined nanostructures, such as DNA origamis, single-stranded tile assemblies and nanogrids. In situ, time-resolved observation reveals that this self-assembly is thermodynamically controlled, proceeds through multiple folding pathways and leads to highly reconfigurable nanostructures. It allows a given system to self-select its most stable shape in a large pool of competitive DNA strands. Strikingly, upon the appearance of a new energy minimum, DNA origamis isothermally shift from one initially stable shape to a radically different one, by massive exchange of their constitutive staple strands. This method expands the repertoire of shapes and functions attainable by isothermal self-assembly and creates a basis for adaptive nanomachines and nanostructure discovery by evolution.
Reference: C. Rossi-Gendron,§ F. El Fakih, § L. Bourdon, K. Nakazawa, J. Finkel, N. Triomphe, L. Chocron, M. Endo, H. Sugiyama, G. Bellot, M. Morel, S. Rudiuk & D. Baigl*. Isothermal self-assembly of multicomponent and evolutive DNA nanostructures. Nat. Nanotechnol. (2023). https://doi.org/10.1038/s41565-023-01468-2
§: equal contribution ; * : correspondence to: damien.baigl@ens.psl.eu
Contact: Damien Baigl (damien.baigl@ens.psl.eu)
