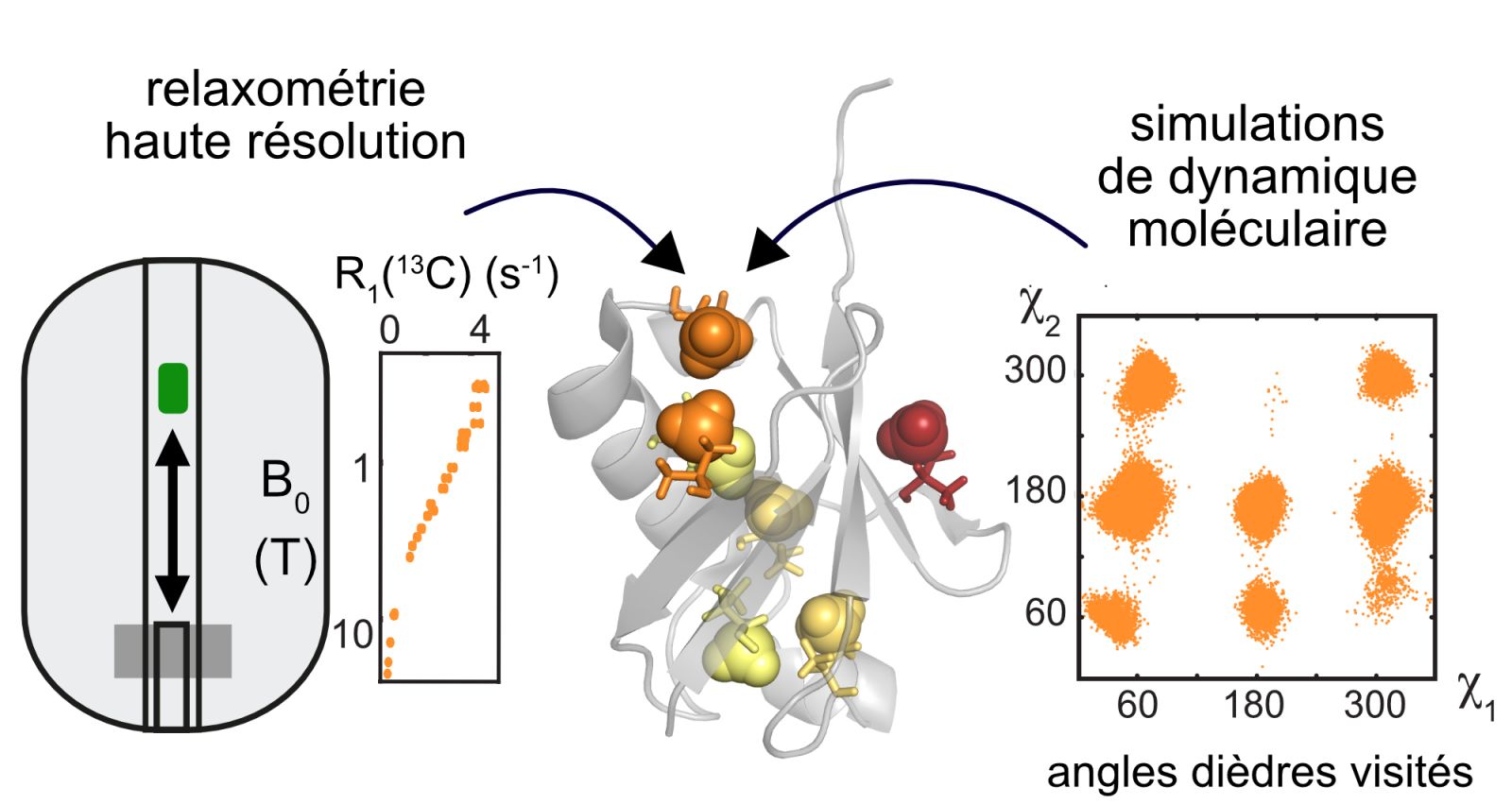
Motions of proteins are essential for the performance of their functions. Aliphatic protein side chains and their motions play critical roles in protein interactions: for recognition and binding of partner molecules at the surface or serving as an entropy reservoir within the hydrophobic core. Here, we present a new NMR method based on highresolution relaxometry and high-fi eld relaxation to determine quantitatively both motional amplitudes and time scales of methyl-bearing side chains in the picosecond-to-nanosecond range. We detect a wide variety of motions in isoleucine side chains in the protein ubiquitin. We unambiguously identify slow motions in the low nanosecond range, which, in conjunction with molecular dynamics computer simulations, could be assigned to transitions between rotamers. Our approach provides unmatched detailed insight into the motions of aliphatic side chains in proteins and provides a better understanding of the nature and functional role of protein side-chain motions.

N’hésitez pas à consulter le communiqué de presse associé à cet article :
Mouvements des protéines : la valse des chaînes latérales
References:
Time-Resolved Protein Side-Chain Motions Unraveled by High-Resolution Relaxometry and Molecular Dynamics Simulations
Samuel F. Cousin, Pavel Kadeřávek, Nicolas Bolik-Coulon, Yina Gu, Cyril Charlier, Ludovic Carlier, Lei Bruschweiler-Li, Thorsten Marquardsen, Jean-Max Tyburn, Rafael Brüschweiler and Fabien Ferrage
Journal of the American Chemical Society, Volume 140, p. 13456 – 13465, 2018
DOI :10.1021/jacs.8b09107
Time-Resolved Protein Side-Chain Motions Unraveled by High-Resolution Relaxometry and Molecular Dynamics Simulations
